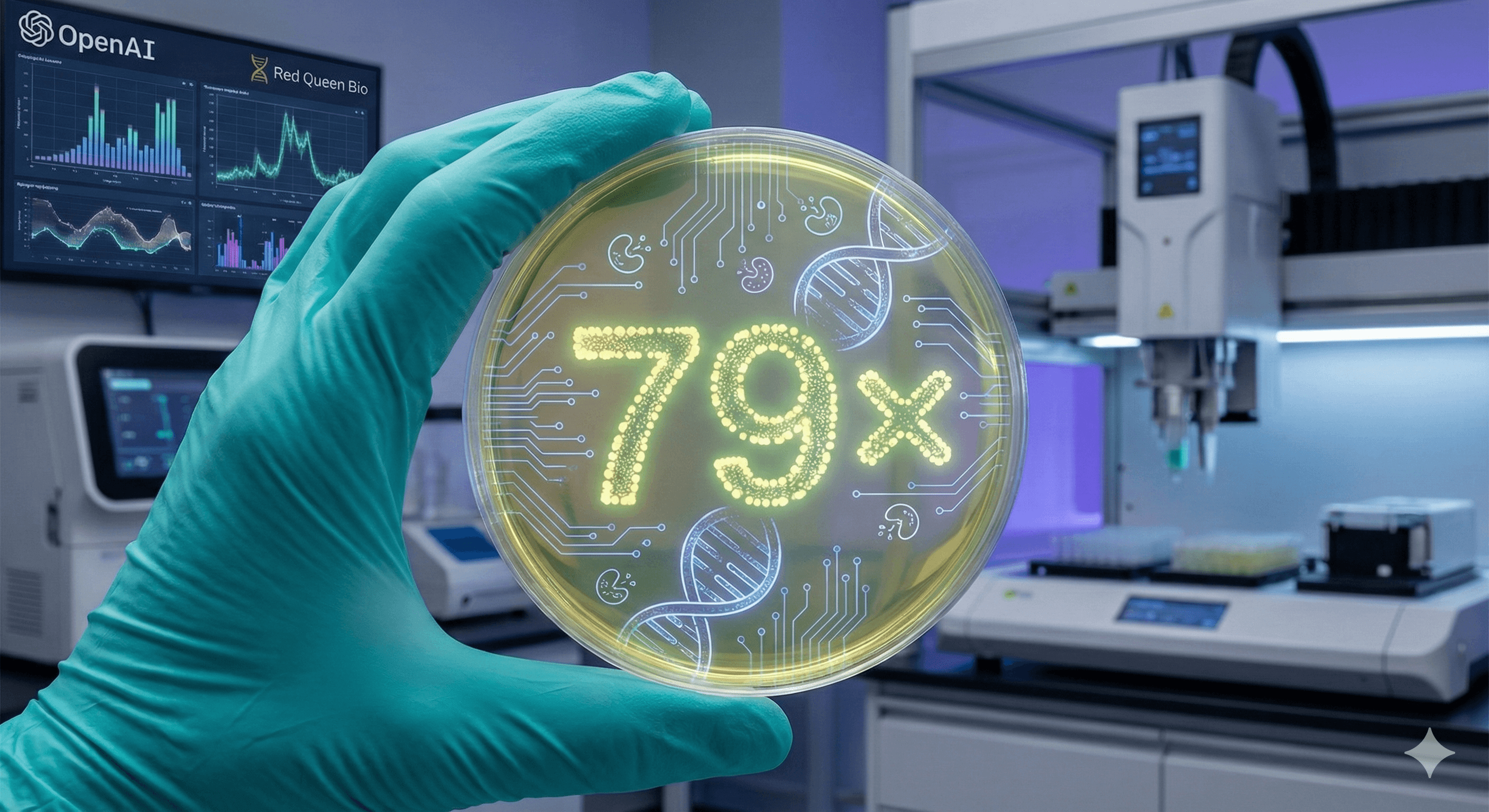
La IA aumenta la eficiencia del clonaje en laboratorios húmedos 79× (OpenAI GPT-5)
La IA aumenta la eficiencia del clonaje en laboratorios húmedos 79× (OpenAI GPT-5)
OpenAI
11 dic 2025

Not sure what to do next with AI?
Assess readiness, risk, and priorities in under an hour.
Not sure what to do next with AI?
Assess readiness, risk, and priorities in under an hour.
➔ Reserva una Consulta
OpenAI informa que GPT-5 mejoró la eficiencia de un protocolo estándar de clonación molecular en un 79× en un estudio de laboratorio húmedo controlado con Red Queen Bio. El modelo propuso cambios novedosos (incluyendo un enfoque de ensamblaje asistido por enzimas) y un ajuste de transformación aparte; los humanos ejecutaron los experimentos y validaron resultados en varias réplicas. Temprano pero significativo.
¿Qué pasó?
OpenAI trabajó con Red Queen Bio para probar si un modelo avanzado podía mejorar significativamente un experimento real. GPT-5 propuso cambios en el protocolo; los científicos realizaron los experimentos y retroalimentaron los resultados; el sistema iteró. Resultado: 79× más clones verificados por secuencia a partir del mismo ADN de entrada en comparación con el método de referencia—la definición del estudio de “eficiencia”.
Por qué importa: la clonación es una piedra angular en la ingeniería de proteínas, pantallas genéticas y la ingeniería de cepas, por lo que un mayor rendimiento por entrada puede acortar ciclos y reducir costos en muchos aspectos de la biología cotidiana.
¿Qué cambió realmente?
Nuevo mecanismo de ensamblaje: GPT-5 sugirió una variación asistida por enzimas que añade dos proteínas auxiliares (RecA y gp32) para mejorar cómo los extremos del ADN se encuentran y emparejan, un paso que limita muchos ensamblajes basados en homología. Solo esto mejoró la eficiencia en el estudio.
Un ajuste separado en la transformación: También propuso un cambio en el manejo durante la transformación que incrementó el número de colonias obtenidas. Juntos, el cambio de ensamblaje y el cambio de transformación entregaron la mejora de 79× de extremo a extremo en las validaciones del estudio.
Nota: El equipo enfatiza que esto se realizó en un sistema benigno, con controles de seguridad estrictos, y que los resultados son tempranos y específicos del sistema—prometedores, pero no una garantía general.
Cómo se midió la “eficiencia 79×”
La eficiencia aquí significa clones verificados por secuencia recuperados por una cantidad fija de ADN de entrada en comparación con el protocolo de clonación de referencia. OpenAI reporta validación en réplicas independientes (n=3) para los principales candidatos.
Lo que esto no significa
No significa que una IA no supervisada esté ejecutando un laboratorio libremente. Los humanos ejecutaron los experimentos; el modelo propuso e iteró.
No significa que la mejora se aplica a todos los organismos, vectores, insertos o flujos de trabajo. El equipo destaca que las ganancias fueron específicas de su configuración y que una generalización más amplia requiere más trabajo.
No elimina preocupaciones de seguridad. El trabajo siguió un marco de preparación y un sistema benigno y restringido para gestionar el riesgo de bioseguridad.
Lo que es genuinamente nuevo
Idea novedosa y mecánicamente fundamentada: el enfoque RecA/gp32 formaliza un paso de “emparejamiento asistido por auxiliares” dentro de un flujo de trabajo estilo Gibson — interesante porque Gibson ha sido un estándar de un solo tubo y una sola temperatura desde 2009.
Evidencia del bucle AI–lab: instrucciones fijas, sin intervención humana en la etapa de propuesta, pero aún así generó un nuevo mecanismo además de una mejora práctica en la transformación.
Señal temprana de robótica: el equipo también ensayó un robot de laboratorio de propósito general que ejecutó protocolos generados por IA; el rendimiento relativo se comparó con experimentos realizados por humanos, aunque con menores rendimientos absolutos (quedan áreas para calibración).
Implicaciones prácticas para líderes de I+D
Esperar ciclos de diseño-fabricación-prueba más rápidos donde se utilizan “sistemas modelo” benignos para desarrollo de métodos, luego adaptados por expertos en el campo.
Planificar la gobernanza: tratar la IA como un motor de propuestas dentro de un marco de seguridad ante todo (revisión de riesgos, control de cambios, historial de auditoría).
Tesis de inversión: si incluso una fracción de estas ganancias se generaliza, el costo/tiempo por paso de clonación podría disminuir materialmente—compuesto a través de la construcción de bibliotecas y programas de detección. La cobertura independiente refleja este potencial pero advierte contra el exceso de publicidad.
Preguntas frecuentes
¿Cómo logró GPT-5 el 79×?
Combinando un nuevo mecanismo de ensamblaje (con proteínas auxiliares para mejorar el emparejamiento) y un cambio en la etapa de transformación, validado en comparación con una base estándar; la métrica fue clones verificados por entrada de ADN fija. OpenAI
¿La IA estaba ejecutando el laboratorio?
No. GPT-5 propuso e iteró; científicos capacitados ejecutaron y subieron los resultados. El estudio utilizó deliberadamente instrucciones fijas para medir las propias contribuciones del modelo. OpenAI
¿Es seguro esto?
Los experimentos se realizaron en un sistema benigno bajo estrictos controles y enmarcados dentro del enfoque de preparación de OpenAI. Los autores destacan explícitamente las consideraciones de bioseguridad. OpenAI
¿Aparecerán las mismas ganancias en mi laboratorio?
No garantizadas. El equipo enfatiza resultados específicos del sistema y estado temprano; se requieren replicación y una evaluación comparativa más amplia. Periodistas independientes también notan la historia del campo de sobreexageración—se aplica un escepticismo saludable. OpenAI
¿Cuál fue la base de referencia?
Un flujo de trabajo de ensamblaje estilo Gibson—ampliamente utilizado para unir fragmentos de ADN. El estudio posiciona sus cambios en relación a esa base. OpenAI
OpenAI informa que GPT-5 mejoró la eficiencia de un protocolo estándar de clonación molecular en un 79× en un estudio de laboratorio húmedo controlado con Red Queen Bio. El modelo propuso cambios novedosos (incluyendo un enfoque de ensamblaje asistido por enzimas) y un ajuste de transformación aparte; los humanos ejecutaron los experimentos y validaron resultados en varias réplicas. Temprano pero significativo.
¿Qué pasó?
OpenAI trabajó con Red Queen Bio para probar si un modelo avanzado podía mejorar significativamente un experimento real. GPT-5 propuso cambios en el protocolo; los científicos realizaron los experimentos y retroalimentaron los resultados; el sistema iteró. Resultado: 79× más clones verificados por secuencia a partir del mismo ADN de entrada en comparación con el método de referencia—la definición del estudio de “eficiencia”.
Por qué importa: la clonación es una piedra angular en la ingeniería de proteínas, pantallas genéticas y la ingeniería de cepas, por lo que un mayor rendimiento por entrada puede acortar ciclos y reducir costos en muchos aspectos de la biología cotidiana.
¿Qué cambió realmente?
Nuevo mecanismo de ensamblaje: GPT-5 sugirió una variación asistida por enzimas que añade dos proteínas auxiliares (RecA y gp32) para mejorar cómo los extremos del ADN se encuentran y emparejan, un paso que limita muchos ensamblajes basados en homología. Solo esto mejoró la eficiencia en el estudio.
Un ajuste separado en la transformación: También propuso un cambio en el manejo durante la transformación que incrementó el número de colonias obtenidas. Juntos, el cambio de ensamblaje y el cambio de transformación entregaron la mejora de 79× de extremo a extremo en las validaciones del estudio.
Nota: El equipo enfatiza que esto se realizó en un sistema benigno, con controles de seguridad estrictos, y que los resultados son tempranos y específicos del sistema—prometedores, pero no una garantía general.
Cómo se midió la “eficiencia 79×”
La eficiencia aquí significa clones verificados por secuencia recuperados por una cantidad fija de ADN de entrada en comparación con el protocolo de clonación de referencia. OpenAI reporta validación en réplicas independientes (n=3) para los principales candidatos.
Lo que esto no significa
No significa que una IA no supervisada esté ejecutando un laboratorio libremente. Los humanos ejecutaron los experimentos; el modelo propuso e iteró.
No significa que la mejora se aplica a todos los organismos, vectores, insertos o flujos de trabajo. El equipo destaca que las ganancias fueron específicas de su configuración y que una generalización más amplia requiere más trabajo.
No elimina preocupaciones de seguridad. El trabajo siguió un marco de preparación y un sistema benigno y restringido para gestionar el riesgo de bioseguridad.
Lo que es genuinamente nuevo
Idea novedosa y mecánicamente fundamentada: el enfoque RecA/gp32 formaliza un paso de “emparejamiento asistido por auxiliares” dentro de un flujo de trabajo estilo Gibson — interesante porque Gibson ha sido un estándar de un solo tubo y una sola temperatura desde 2009.
Evidencia del bucle AI–lab: instrucciones fijas, sin intervención humana en la etapa de propuesta, pero aún así generó un nuevo mecanismo además de una mejora práctica en la transformación.
Señal temprana de robótica: el equipo también ensayó un robot de laboratorio de propósito general que ejecutó protocolos generados por IA; el rendimiento relativo se comparó con experimentos realizados por humanos, aunque con menores rendimientos absolutos (quedan áreas para calibración).
Implicaciones prácticas para líderes de I+D
Esperar ciclos de diseño-fabricación-prueba más rápidos donde se utilizan “sistemas modelo” benignos para desarrollo de métodos, luego adaptados por expertos en el campo.
Planificar la gobernanza: tratar la IA como un motor de propuestas dentro de un marco de seguridad ante todo (revisión de riesgos, control de cambios, historial de auditoría).
Tesis de inversión: si incluso una fracción de estas ganancias se generaliza, el costo/tiempo por paso de clonación podría disminuir materialmente—compuesto a través de la construcción de bibliotecas y programas de detección. La cobertura independiente refleja este potencial pero advierte contra el exceso de publicidad.
Preguntas frecuentes
¿Cómo logró GPT-5 el 79×?
Combinando un nuevo mecanismo de ensamblaje (con proteínas auxiliares para mejorar el emparejamiento) y un cambio en la etapa de transformación, validado en comparación con una base estándar; la métrica fue clones verificados por entrada de ADN fija. OpenAI
¿La IA estaba ejecutando el laboratorio?
No. GPT-5 propuso e iteró; científicos capacitados ejecutaron y subieron los resultados. El estudio utilizó deliberadamente instrucciones fijas para medir las propias contribuciones del modelo. OpenAI
¿Es seguro esto?
Los experimentos se realizaron en un sistema benigno bajo estrictos controles y enmarcados dentro del enfoque de preparación de OpenAI. Los autores destacan explícitamente las consideraciones de bioseguridad. OpenAI
¿Aparecerán las mismas ganancias en mi laboratorio?
No garantizadas. El equipo enfatiza resultados específicos del sistema y estado temprano; se requieren replicación y una evaluación comparativa más amplia. Periodistas independientes también notan la historia del campo de sobreexageración—se aplica un escepticismo saludable. OpenAI
¿Cuál fue la base de referencia?
Un flujo de trabajo de ensamblaje estilo Gibson—ampliamente utilizado para unir fragmentos de ADN. El estudio posiciona sus cambios en relación a esa base. OpenAI
Recibe consejos prácticos directamente en tu bandeja de entrada
Al suscribirte, das tu consentimiento para que Generation Digital almacene y procese tus datos de acuerdo con nuestra política de privacidad. Puedes leer la política completa en gend.co/privacy.
Generación
Digital

Oficina en el Reino Unido
33 Queen St,
Londres
EC4R 1AP
Reino Unido
Oficina en Canadá
1 University Ave,
Toronto,
ON M5J 1T1,
Canadá
Oficina NAMER
77 Sands St,
Brooklyn,
NY 11201,
Estados Unidos
Oficina EMEA
Calle Charlemont, Saint Kevin's, Dublín,
D02 VN88,
Irlanda
Oficina en Medio Oriente
6994 Alsharq 3890,
An Narjis,
Riyadh 13343,
Arabia Saudita
Número de la empresa: 256 9431 77 | Derechos de autor 2026 | Términos y Condiciones | Política de Privacidad
Generación
Digital

Oficina en el Reino Unido
33 Queen St,
Londres
EC4R 1AP
Reino Unido
Oficina en Canadá
1 University Ave,
Toronto,
ON M5J 1T1,
Canadá
Oficina NAMER
77 Sands St,
Brooklyn,
NY 11201,
Estados Unidos
Oficina EMEA
Calle Charlemont, Saint Kevin's, Dublín,
D02 VN88,
Irlanda
Oficina en Medio Oriente
6994 Alsharq 3890,
An Narjis,
Riyadh 13343,
Arabia Saudita