AI Enhances Wet-Lab Cloning Efficiency by 79 Times (OpenAI GPT-5)
AI Enhances Wet-Lab Cloning Efficiency by 79 Times (OpenAI GPT-5)
OpenAI
Dec 11, 2025

Not sure what to do next with AI?
Assess readiness, risk, and priorities in under an hour.
Not sure what to do next with AI?
Assess readiness, risk, and priorities in under an hour.
➔ Schedule a Consultation
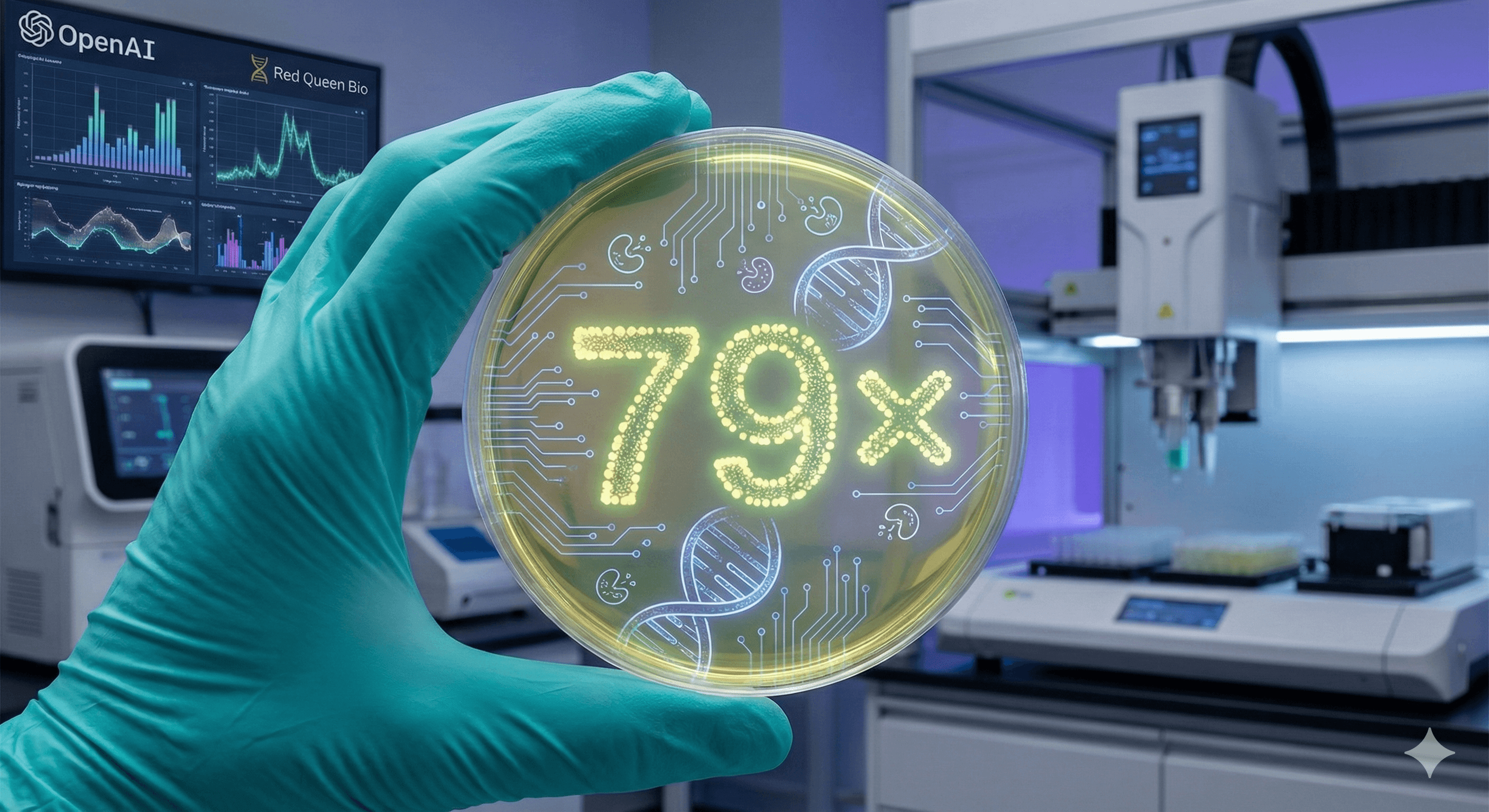
OpenAI has announced that GPT-5 enhanced the efficiency of a standard molecular cloning protocol by 79× in a controlled wet-lab study with Red Queen Bio. The model suggested novel changes, including an enzyme-assisted assembly approach and an independent transformation adjustment; humans conducted the experiments and confirmed results across replicates. These are early yet significant findings.
What happened?
OpenAI partnered with Red Queen Bio to assess if an advanced model could significantly improve a real-world experiment. GPT-5 proposed protocol modifications; scientists performed the experiments and fed back the results; the system iterated. Outcome: 79× more sequence-verified clones from the same DNA input compared to the baseline method — this was the study’s measure of “efficiency”.
Why it matters: Cloning is crucial across protein engineering, genetic screens, and strain engineering, so a higher yield per input can shorten cycles and reduce costs across numerous everyday biological applications.
What actually changed?
New assembly mechanism: GPT-5 suggested an enzyme-assisted variation that introduces two helper proteins (RecA and gp32) to improve DNA end pairing — a step that constrains many homology-based assemblies. This alone improved efficiency in the study.
A separate transformation tweak: The model also proposed a procedural change during transformation that increased the number of colonies obtained. Together, the assembly and transformation changes delivered the 79× improvement in the study’s validation runs.
Note: The team highlights that this was performed in a benign system, with strict safety controls, emphasizing that these are early-stage and system-specific results—promising, but not a universal assurance.
How “79× efficiency” was measured
Efficiency in this context refers to sequence-verified clones recovered per set amount of input DNA compared to the baseline cloning protocol. OpenAI reports validation across independent replicates (n=3) for top candidates.
What this doesn’t mean
It doesn’t mean unsupervised AI is running a free-form lab. Human experts executed the experiments; the model proposed and iterated.
It doesn’t mean the improvement is applicable to every organism, vector, insert, or workflow. The team notes that the gains were specific to their setup and that broader generalization requires additional work.
It doesn’t eliminate safety concerns. The research adhered to a preparedness framework and a constrained, benign system to manage biosecurity risks.
What’s genuinely new
Novel, mechanism-based idea: The RecA/gp32 approach formalizes a “helper-assisted pairing” step within a Gibson-style workflow — notable since Gibson has been a one-tube, one-temperature method since 2009.
AI–lab loop evidence: Fixed prompting, with no human direction during the proposal stage, yet it still led to a new mechanism plus a practical transformation improvement.
Early robotics signal: The team also tried a general-purpose lab robot running AI-generated protocols; relative performance tracked human-run experiments, though with lower absolute yields (areas for calibration remain).
Practical implications for R&D leaders
Expect faster design–make–test cycles using benign “model systems” for method development, then adaptation by domain experts.
Plan governance: Consider AI as a proposal tool within a safety-first framework (risk review, change control, audit trail).
Investment thesis: If even a fraction of these gains generalize, the cost/time per cloning step could significantly decrease — compounding across library construction and screening programs. Independent reviews echo this potential but advise caution against excessive optimism.
FAQs
How did GPT-5 achieve 79×?
By employing a new assembly mechanism (using helper proteins to improve pairing) and a transformation-stage change, validated against a standard baseline; the metric was verified clones per fixed DNA input. OpenAI
Was the AI running the lab?
No. GPT-5 suggested and iterated proposals; skilled scientists conducted and reported results. The study used fixed prompts intentionally to quantify the model’s contributions. OpenAI
Is this safe?
The experiments took place in a benign system under strict controls, framed within OpenAI’s preparedness strategy. The authors explicitly consider biosecurity implications. OpenAI
Will the same gains appear in my lab?
Not guaranteed. The team emphasizes system-specific results and the early-stage nature; further replication and broader benchmarking are necessary. Independent journalists also point out the field’s history of overstatements — maintain healthy skepticism. OpenAI
What was the baseline?
A Gibson-style assembly workflow — widely employed for joining DNA fragments. The study contextualizes its changes relative to this baseline. OpenAI
OpenAI has announced that GPT-5 enhanced the efficiency of a standard molecular cloning protocol by 79× in a controlled wet-lab study with Red Queen Bio. The model suggested novel changes, including an enzyme-assisted assembly approach and an independent transformation adjustment; humans conducted the experiments and confirmed results across replicates. These are early yet significant findings.
What happened?
OpenAI partnered with Red Queen Bio to assess if an advanced model could significantly improve a real-world experiment. GPT-5 proposed protocol modifications; scientists performed the experiments and fed back the results; the system iterated. Outcome: 79× more sequence-verified clones from the same DNA input compared to the baseline method — this was the study’s measure of “efficiency”.
Why it matters: Cloning is crucial across protein engineering, genetic screens, and strain engineering, so a higher yield per input can shorten cycles and reduce costs across numerous everyday biological applications.
What actually changed?
New assembly mechanism: GPT-5 suggested an enzyme-assisted variation that introduces two helper proteins (RecA and gp32) to improve DNA end pairing — a step that constrains many homology-based assemblies. This alone improved efficiency in the study.
A separate transformation tweak: The model also proposed a procedural change during transformation that increased the number of colonies obtained. Together, the assembly and transformation changes delivered the 79× improvement in the study’s validation runs.
Note: The team highlights that this was performed in a benign system, with strict safety controls, emphasizing that these are early-stage and system-specific results—promising, but not a universal assurance.
How “79× efficiency” was measured
Efficiency in this context refers to sequence-verified clones recovered per set amount of input DNA compared to the baseline cloning protocol. OpenAI reports validation across independent replicates (n=3) for top candidates.
What this doesn’t mean
It doesn’t mean unsupervised AI is running a free-form lab. Human experts executed the experiments; the model proposed and iterated.
It doesn’t mean the improvement is applicable to every organism, vector, insert, or workflow. The team notes that the gains were specific to their setup and that broader generalization requires additional work.
It doesn’t eliminate safety concerns. The research adhered to a preparedness framework and a constrained, benign system to manage biosecurity risks.
What’s genuinely new
Novel, mechanism-based idea: The RecA/gp32 approach formalizes a “helper-assisted pairing” step within a Gibson-style workflow — notable since Gibson has been a one-tube, one-temperature method since 2009.
AI–lab loop evidence: Fixed prompting, with no human direction during the proposal stage, yet it still led to a new mechanism plus a practical transformation improvement.
Early robotics signal: The team also tried a general-purpose lab robot running AI-generated protocols; relative performance tracked human-run experiments, though with lower absolute yields (areas for calibration remain).
Practical implications for R&D leaders
Expect faster design–make–test cycles using benign “model systems” for method development, then adaptation by domain experts.
Plan governance: Consider AI as a proposal tool within a safety-first framework (risk review, change control, audit trail).
Investment thesis: If even a fraction of these gains generalize, the cost/time per cloning step could significantly decrease — compounding across library construction and screening programs. Independent reviews echo this potential but advise caution against excessive optimism.
FAQs
How did GPT-5 achieve 79×?
By employing a new assembly mechanism (using helper proteins to improve pairing) and a transformation-stage change, validated against a standard baseline; the metric was verified clones per fixed DNA input. OpenAI
Was the AI running the lab?
No. GPT-5 suggested and iterated proposals; skilled scientists conducted and reported results. The study used fixed prompts intentionally to quantify the model’s contributions. OpenAI
Is this safe?
The experiments took place in a benign system under strict controls, framed within OpenAI’s preparedness strategy. The authors explicitly consider biosecurity implications. OpenAI
Will the same gains appear in my lab?
Not guaranteed. The team emphasizes system-specific results and the early-stage nature; further replication and broader benchmarking are necessary. Independent journalists also point out the field’s history of overstatements — maintain healthy skepticism. OpenAI
What was the baseline?
A Gibson-style assembly workflow — widely employed for joining DNA fragments. The study contextualizes its changes relative to this baseline. OpenAI
Receive practical advice directly in your inbox
By subscribing, you agree to allow Generation Digital to store and process your information according to our privacy policy. You can review the full policy at gend.co/privacy.
Generation
Digital

Business Number: 256 9431 77 | Copyright 2026 | Terms and Conditions | Privacy Policy
Generation
Digital